Sapientia
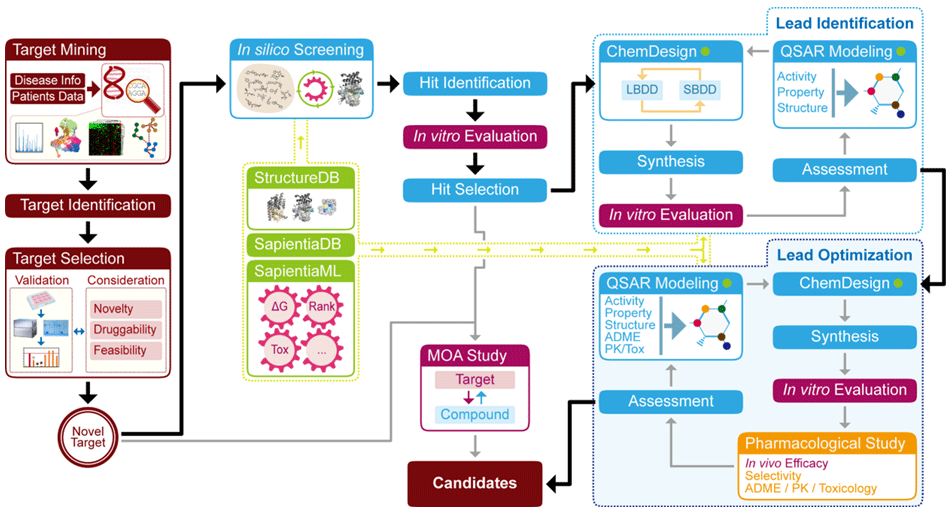
Sapientia is a well-organized drug discovery platform which integrates machine learning (ML) technology with wet lab experimentation, facilitating drastic acceleration of drug discovery from target identification to selection of a lead candidate [Figure 1: WORKFLOW#1]. Domain of in silico analysis (the left circuit of the diagram) serves a virtual provider of either a druggable gene target(s) or a new chemical hit/lead(s) to expand a pool of source for therapeutically promising and reasonable ingredients. Domain of experimentation (the right circuit of the diagram) plays a front role as a validator to investigate whether the designed (or suggested) compounds may have an effectiveness on the disease models in aspect of medicinal chemistry and biology. Several contact points of two developmental circuits such as QSAR (quantitative structure-activity relationship) and chemical synthesis actively communicate with each other to sustain high probability of success.
Sapientia operates through four core parts: (1) genomic workflow from target mining to selection of a new druggable gene, (2) hit identification and selection for the novel target gene, (3) integrated workflow for compound design and synthesis for lead identification and optimization, and (4) SapientiaCore, a bunch of computational resources such as StructureDB, SapientiaDB and SapientiaML
The first core of Sapientia called SapientiaTID identifies a novel druggable gene(s) for development of a first-in-class drug. Generally, the drug discovery and development begin with target identification and characterization. Successful drug discovery and development depends on how much we know the clinical spectrum of disease and which gene(s) is a potentially ‘druggable’ therapeutic target. Most of targets (i.e., genes) for use of drug discovery are usually identified from scientific literature and publicly available databases. Otherwise, either target deconvolution–a way to identify a target if a biologically effective compound already exists–or target discovery via high throughput screening at excavation of a new druggable target can be a second option. The target identification and validation (TID) facility of Sapientia is launched to achieve successful and efficient target discovery via genomic and experimental approaches. Genomic data obtained from patients, model animals, and cell cultures via single-cell RNA sequencing (scRNA-seq), total RNA sequencing (RNA-seq), genome-wide association study (GWAS), etc. were multifacetedly analyzed through statistical approaches to understand the routine of pathogenesis and to suggest a set of causative genes. TID also attempts to empirically validate the confidence of suggested genes via experimental methodology, specifically silencing-based assay with known model systems (e.g., usefulness of normal lung fibroblasts as an experimental model of pulmonary fibrosis). During or after the validation, a final gene(s) of interests called as a NOVEL TARGET is selected under three critical points: (1) novelty, (2) draggability, and (3) feasibility

The step for hit identification and selection designated as SapientiaHITS is initiated by versatile strategy of Sapientia’s in silico screening supported by SapientiaCORE, a cooperative combination of conventional computer-aided drug discovery (CADD) such as ligand-based (LB) and structure-based (SB) drug discovery (DD) [LBDD and SBDD, respectively] with StructureDB, SapientiaDB and SapientiaML. Particularly, SapientiaDB, the virtual informative reservoir of compounds which are derived from in-house design and synthesis as well as publicly available databases such as ChEMBL, PubChem, ZINC, Mcule, etc. provides the diversity of pools with the initial screening step. Furthermore, SapientiaML effectively and accurately support the virtual process of hit identification via high-performance of statistical models such as CNN (convolutive neural network), RNN (recursive neural network) and so on. A line of hit compounds identified by SapientiaCORE are experimentally evaluated to see whether they are effective worthy to be considered as a starting material for the next discovery step.
SapientiaLEAD and SapientiaOPT has the responsibilities to promote lead identification and optimization by interdisciplinary interwining chemistry design to synthesis to biological validation, respectively. Most critical capability in two modules is that ChemDesign, a unification of CADD with statistical ML approaches, can produce the diverse series of novel effective compounds which have never been published elsewhere even though the three-dimensional structure of a target is never known. Our ChemDesign can generate the variety of reasonable scaffolds via our ML approaches. Furthermore, Sapientia’s in-house facility to support fast synthesis of compounds permits to speed up the design-validation cycle. And our computational strategies for QSAR modelling help us decide whether a compound could jump to the next step.
